RNA preprocessing#
Method#
FastQC was used before and after trimming to check the quality of RNA short reads. Trimmomatic was used to trim the reads.
FastQC#
Command:
for file in ~/genome_analysis/data/RNA_raw_data/*
do
fastqc $file -o ~/genome_analysis/analyses/04_rna_preprocessing/01-fastqc
done
for file in ~/genome_analysis/data/RNA_trimmed_reads/*
do
fastqc $file -o ~/genome_analysis/analyses/04_rna_preprocessing/03-fastqc
done
Trimmomatic#
Command:
java -jar $TRIMMOMATIC_HOME/trimmomatic.jar PE -threads 2 \
~/genome_analysis/data/RNA_raw_data/${name}_1.fastq.gz \
~/genome_analysis/data/RNA_raw_data/${name}_2.fastq.gz \
~/genome_analysis/analyses/04_rna_preprocessing/02-trimmomatic/$name.1P.fastq.gz \
~/genome_analysis/analyses/04_rna_preprocessing/02-trimmomatic/$name.1U.fastq.gz \
~/genome_analysis/analyses/04_rna_preprocessing/02-trimmomatic/$name.2P.fastq.gz \
~/genome_analysis/analyses/04_rna_preprocessing/02-trimmomatic/$name.2U.fastq.gz \
ILLUMINACLIP:$TRIMMOMATIC_HOME/adapters/TruSeq3-PE.fa:2:30:10 \
LEADING:20 \
TRAILING:20 \
SLIDINGWINDOW:1:3 \
MINLEN:40 \
MAXINFO:40:0.5
In paired end mode, two input files, and 4 output files are specified, 2 for the ‘paired’ output where both reads survived the processing, and 2 for corresponding ‘unpaired’ output where a read survived, but the partner read did not.
Options were set to be the same as the paper.
Results and discussion#
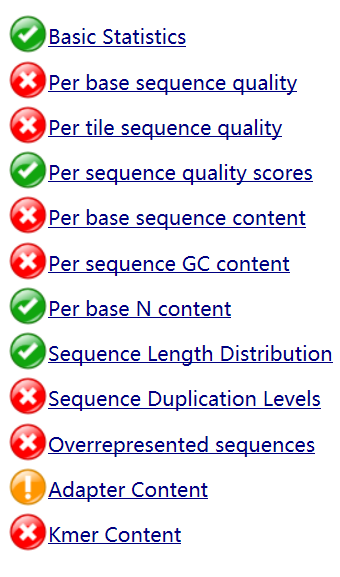
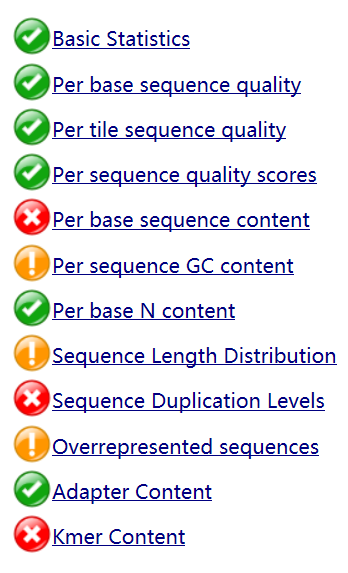
The main purpose of trimming is to remove adapters which will lead to mapping errors because adapter sequences do not occur in genome. Take one of the file ERR2036629 as an example. The quality before trimming is low, while quality clearly improves after trimming.
Input Read Pairs: 24278349
Both Surviving: 21793091 (89.76%)
Forward Only Surviving: 2433801 (10.02%)
Reverse Only Surviving: 10914 (0.04%)
Dropped: 40543 (0.17%)
before |
after |
|---|---|
|
|
Q & A#
Reads quality control#
What is the structure of a FASTQ file?
Four lines including a sequence identifier with information about the sequencing run and the cluster, the sequence, a separator a plus sign, the base call quality scores.
How is the quality of the data stored in the FASTQ files? How are paired reads identified?
Quality score for each base is represented as an ASCII character. Two reads in a read pair are store in two seperate files, having the same prefix in the first line, identified as the first or second read by suffix 1 or 2.
How is the quality of your data?
RNAseq reads have low quality before trimming, according to the red crosses on several metrics in the FastQC reports.
What can generate the issues you observe in your data? Can these cause any problems during subsequent analyses?
The existence of the adapter, possible contamination in experiments.
Reads preprocessing#
How many reads have been discarded after trimming?
Less than 1% RNA reads were discarded. In the trimming process of CANU of DNA reads, only 8772 reads survive from 215276 input reads.
How can this affect your future analyses and results?
Trimming RNA reads increases the accuracy of RNA mapping and counting. Most DNA reads deleted are reads with no overlaps, so they have no effect on assembly.
How is the quality of your data after trimming?
Although some metrics still have low quality, total quality is improved.
What do the LEADING, TRAILING and SLIDINGWINDOW options do?
LEADING cuts bases off the start of a read if below a threshold quality. TRAILING cuts bases off the end of a read if below a threshold quality. SLIDINGWINDOW performs a sliding window trimming, cutting once the average quality within the window falls below a threshold. The different processing steps occur in the order in which the steps are specified on the command line.